
- #CLC GENOMICS WORKBENCH 5.5 ARCHIVE#
- #CLC GENOMICS WORKBENCH 5.5 SOFTWARE#
- #CLC GENOMICS WORKBENCH 5.5 DOWNLOAD#
papatasi with 0.84 ☌ Tm and unique curve based on thermodynamic differences was an important criteria using HRM technique.Ĭonclusion: Subsequent war in Iraq made a high risk habitat for parasites transmission. In amplified fragment of COII gene among 611 bp, 452 bp had no genetic variations with low polymorphic sites (P=0.001) and high synonymous (79.8%) as compare to non-synonymous sites (20.2%). Results: Among about 3000 collected sandflies, 89 female P. The melting curve plots were generated and DNA sequences were analyzed using Sequencher 3.1.1, CLC Main Workbench 5.5, MEGA 6, DnaSP5.10.01 and MedCalc® version 13.3.3 soft wares. PCR products cloned into pTG19-T vector, then purified plasmid concentration was measured at 260 and 280 nm wavelength. Methods: Modified and developed method of High Resolution Melting (HRM) as a preferable technique was employed to accurate identification of Leishmania in sandflies from Iranian border with Iraq, by targeting cytochrome oxidase II (COII) gene and designing suitable primers.
#CLC GENOMICS WORKBENCH 5.5 ARCHIVE#
It also includes a migration to version 68 of Ensembl a new variation dataset for Puccinia graminis a structural variation dataset for Sorghum bicolor imported from the database of genomic variants archive and improvements to assembly, annotation and variation datasets among other updates.Background: Firmly identification of Leishmania in Phlebotomus papatasi and understanding of natural transmission cycles of parasites in sandflies are important for treatment and local control. This release adds two new genomes to Ensembl Fungi - Leptosphaeria maculans and Trichoderma virens - bringing the total genomes supported to 354. Also in this version, the water orientation VIDA extension has been completely rewritten to make it easier to use and to find key waters and understand their interactions and several bugs have been fixed. It also produces hypotheses of ligand modifications to improve affinity, based on the energetics of the water environment directly adjacent to the ligand.
The release includes a new command-line program called GamePlan that runs several SZMAP calculations and analyzes the results to examine how the existing ligand chemistry aligns with the pocket environment.
#CLC GENOMICS WORKBENCH 5.5 SOFTWARE#
OpenEye Scientific Software has released SZMAP v1.1.0, the latest version of the company’s software for studying water’s role in molecular interactions. It also improves the assembly of data from Ion Torrent’s PGM and adds a new coverage normalization capability to NextGene’s Floton assembler generates reports automatically and imports and filters data from the Catalogue of Somatic Mutations in Cancer among other updates. The release includes new tools for HLA analysis and accepts the SOLiD XSQ data format. SoftGenetics has also released a new version of its NextGene software, which is used for analyzing next-generation sequence data. Users also have access to applications in the GeneMarker software suite, such as multiplex ligation-dependent probe amplification, trisomy analysis, and more. SoftGenetics has released GeneMarkerMTP, a multi-template processor for use in genotyping applications.Īccording to its developers, GeneMarkerMTP can process varying chemistries or size standards in a single analysis run.
#CLC GENOMICS WORKBENCH 5.5 DOWNLOAD#
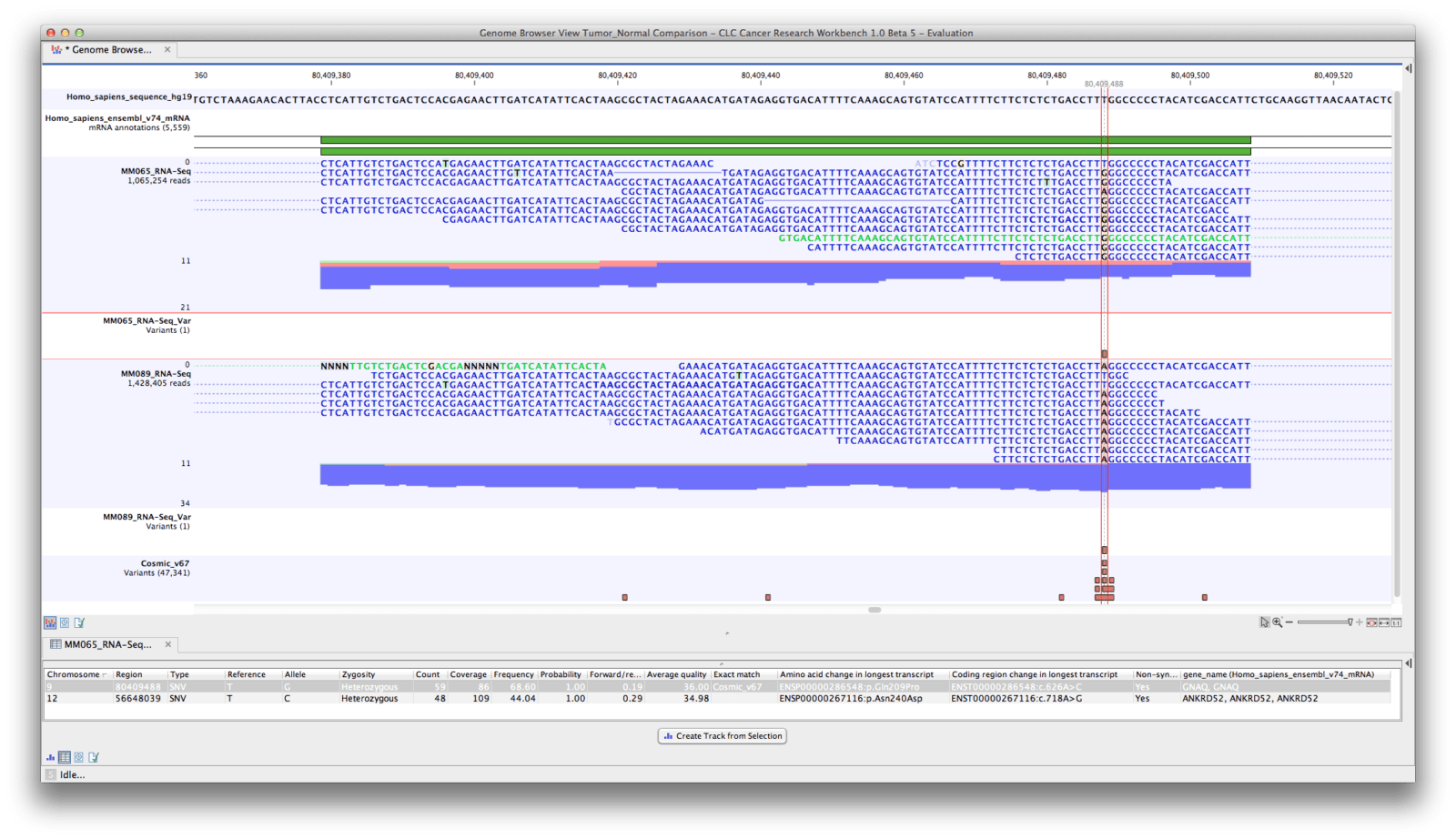
Additionally, users can create workflows that combine various tools from the toolbox into a single analysis step have access to tools for advanced variant detection, which are well suited for genomes of higher ploidy and can easily download genomic sequences and annotation data from public databases among other capabilities. This week, CLC Bio released CLC Genomics Workbench 5.5, the latest version of its desktop software and CLC Genomics Server 4.5, the newest release of its enterprise software.īoth software packages now include tools to build complete resequencing workflows including read mapping, variant detection, and downstream analysis. The latest version of the tool can accept and publish very large images, made up of many individual tiles, and make them viewable online. Glencoe Software and the Journal of Cell Biology have updated the JCB DataViewer, a platform for presenting, sharing, and archiving published scientific image data that is based on technology developed by the Open Microscopy Environment.

Advances in Clinical Genomics Profiling.


 0 kommentar(er)
0 kommentar(er)
